Musing 132: K-Dense Analyst: Towards Fully Automated Scientific Analysis
Interesting paper out of Biostate AI (Palo Alto) and Bayosthiti AI (Bengaluru, India)
Today’s paper: K-Dense Analyst: Towards Fully Automated Scientific Analysis. Li et al. 9 Aug, 2025. https://arxiv.org/pdf/2508.07043
The gap between how quickly life sciences data can be generated and how quickly it can be analyzed has been widening for years. A single genomics run can produce terabytes of raw data, but the downstream analysis with its quality checks, statistical modeling, and integration of pathway databases can take months. This analysis bottleneck has become a major constraint on discovery.
Large language models (LLMs) have shown promise in scientific reasoning, but when confronted with the realities of modern bioinformatics workflows, even the most advanced models stumble. Recent benchmarks confirm this: frontier models that perform well on general reasoning tests achieve less than 23% accuracy on BixBench, a rigorous set of open-ended bioinformatics challenges.
In today’s paper, the authors propose K-Dense Analyst, which approaches the problem differently. It is a hierarchical, multi-agent framework designed for autonomous scientific analysis. Rather than simply prompting a powerful model, it combines two tightly coupled feedback loops, one for planning and one for implementation.
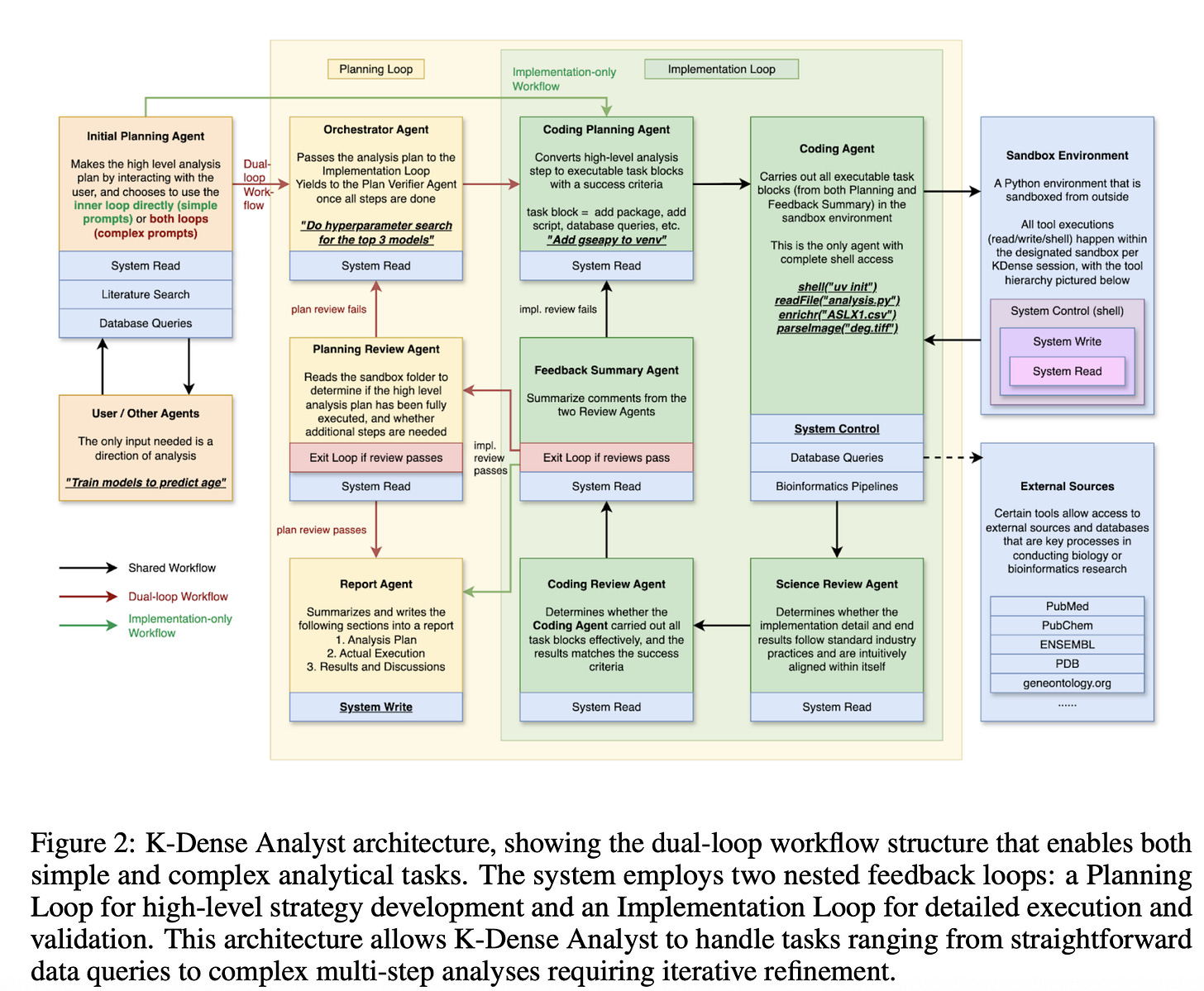
As shown in Figure 2 above, the Planning Loop develops a high-level analytical strategy, deciding whether a problem requires a quick database query or a full differential expression analysis. The Implementation Loop then executes the strategy in a secure sandbox, producing code, running analyses, and validating results at each stage. Ten specialized agents inhabit this architecture, some responsible for breaking down complex objectives into executable steps, others for reviewing the technical correctness of code or assessing the validity of the methodology. The clear separation between strategy and execution, with validation at both levels, allows the system to adapt on the fly, skipping unnecessary steps for simple tasks or iterating until the desired quality is reached for complex ones.
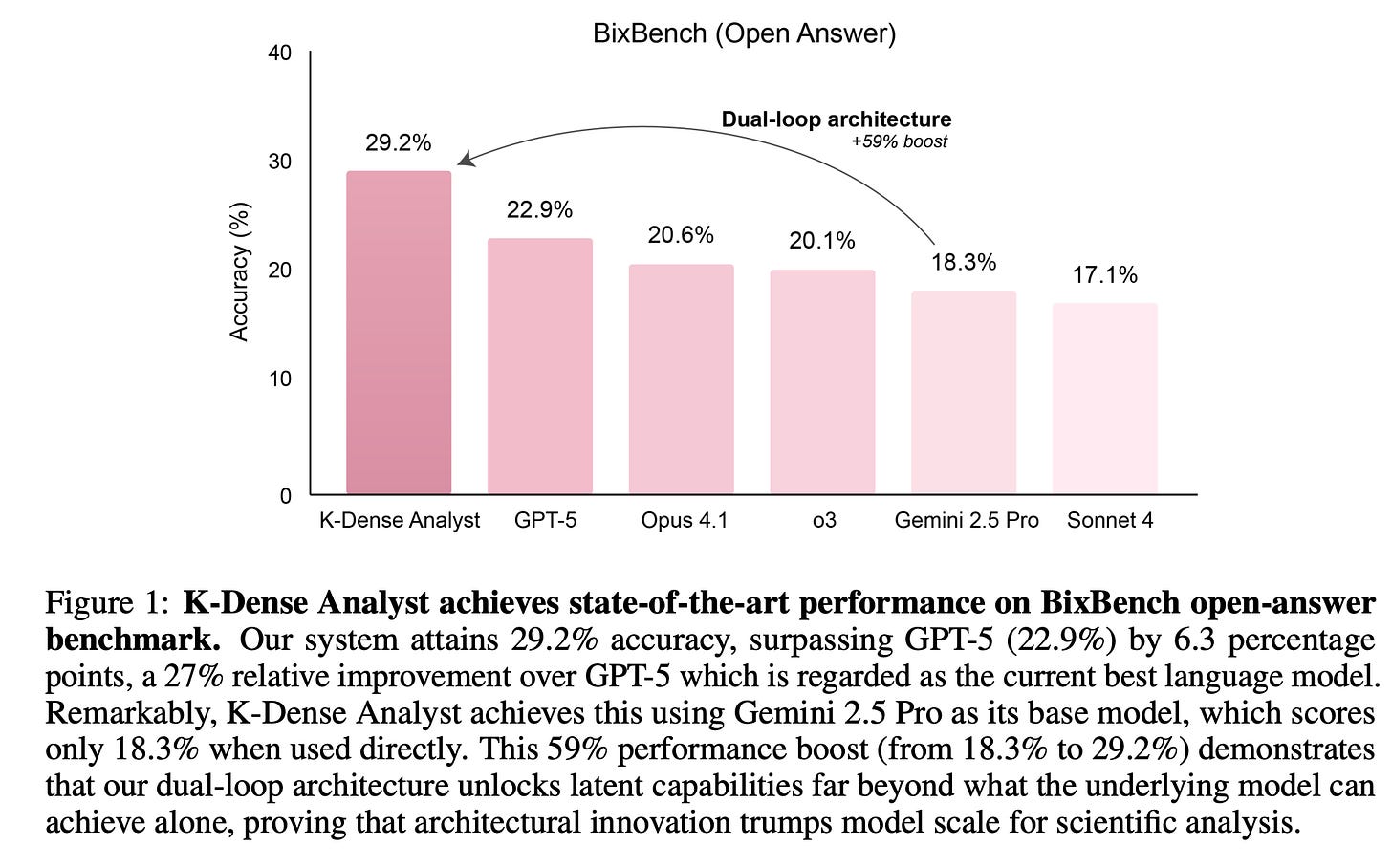
The impact of this architecture is clear in Figure 1 below. On BixBench’s open-answer challenges, K-Dense Analyst scores 29.2 percent accuracy, a 6.3 point improvement over GPT-5 at 22.9 percent and well ahead of models like Opus 4.1 at 20.6 percent or Gemini 2.5 Pro at 18.3 percent. That last number is notable because Gemini 2.5 Pro is the base model inside K-Dense Analyst. Running Gemini alone yields 18.3 percent, but embedding it within this multi-agent framework boosts performance by 59 percent. This suggests that architecture, not just model size, determines success in complex analytical domains.
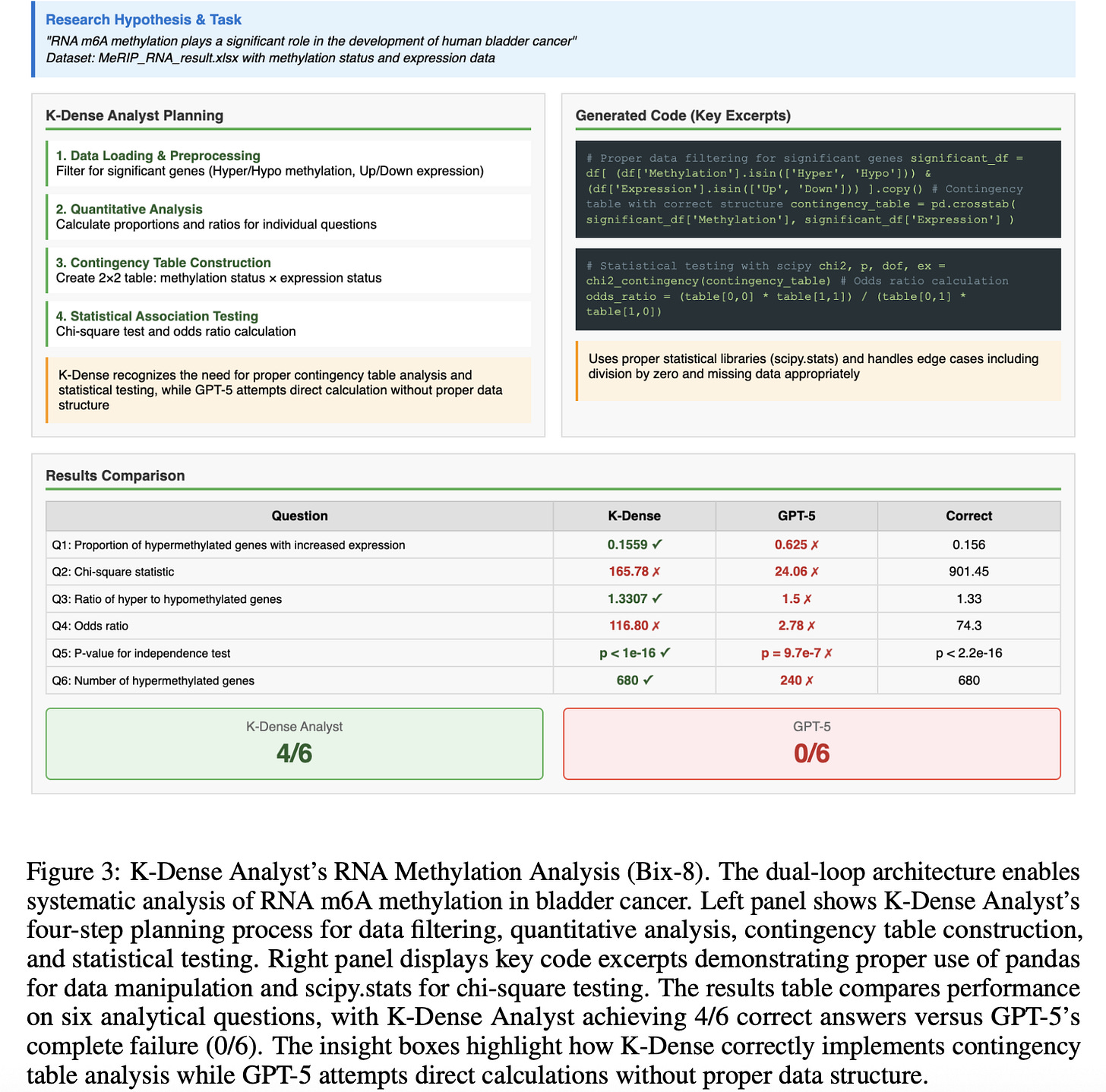
The case study figures below show how this works in practice. In an RNA methylation analysis, K-Dense Analyst planned the correct statistical tests and generated validated code for contingency table construction and chi-square testing, while GPT-5 skipped essential steps and got zero correct answers.
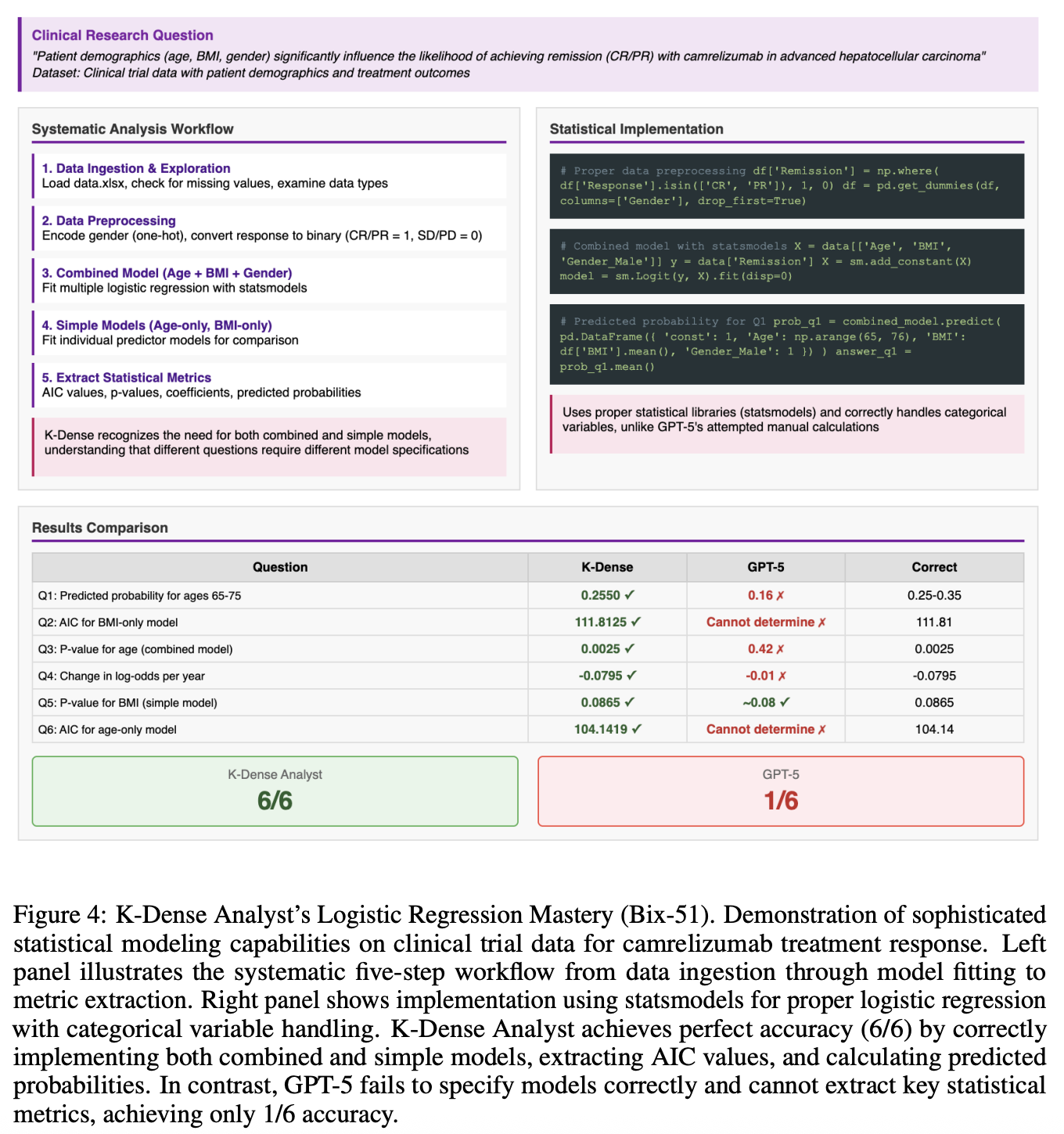
In a logistic regression modeling task, the system built multiple models, extracted AIC values, and interpreted p-values correctly, scoring six out of six, while GPT-5 mis-specified models and misread metrics. Finally, in a challenging microbial co-culture study, K-Dense Analyst implemented proper statistical corrections for Dunnett’s post-hoc tests and calculated phenotypic distance metrics, handling domain-specific computation that a language-only model could not manage.
In closing this musing, I want to note some caveats that are also voiced by the authors. They found at least one mislabeled answer in BixBench, suggesting benchmark quality can distort performance comparisons. The system’s dependence on a closed-source API limits reproducibility, though support for open models like Qwen3 is planned. Tasks involving very recent literature, emerging tools, or fine parameter tuning remain challenging. These are areas where the broader K-Dense platform adds capabilities such as on-demand tool creation, real-time literature search, and continual knowledge integration.
For industry, the message is that operationalizing AI for scientific workflows will require more than better models. Purpose-built agentic frameworks with embedded validation, modular tool integration, and adaptive execution are likely to be necessary. For researchers, the takeaway is that autonomous scientific reasoning will not emerge from scaling LLMs alone. It requires systems that can connect high-level reasoning to low-level computation in a robust and verifiable way. The dual-loop approach demonstrated here could be adapted for other scientific fields such as chemistry, climate science, or structural biology. The principle of separating strategic decomposition from tactical execution, with mandatory validation at each stage, is not limited to bioinformatics. If the aim is to develop autonomous AI co-scientists, K-Dense Analyst may represent less a one-off breakthrough than an early example of the architectures that will make them possible.