Musing 135: Addressing accuracy and hallucination of LLMs in Alzheimer's disease research through knowledge graphs
Excellent paper on knowledge graphs and its application to Alzheimer's research from two academic labs (Washington University in St. Louis and UWashington)
Today’s paper: Addressing accuracy and hallucination of LLMs in Alzheimer's disease research through knowledge graphs. Xu et al. Sept 2, 2025. https://arxiv.org/pdf/2508.21238
The past two years have seen large language models (LLMs) like ChatGPT explode into public awareness. Their ability to generate human-like text and answer questions with fluidity is unlike anything we’ve had before. But ask a deeper, domain-specific question (say, about the kinetics of amyloid beta peptides in Alzheimer’s disease) and the cracks start to show. The model may produce an answer that sounds plausible, but it risks being incomplete, misleading, or worse, fabricated. In biomedicine, where precision and traceability matter, these shortcomings become unacceptable.
This is the challenge that today’s paper tackles head-on. The authors argue that while LLMs hold great promise for scientific use, they must be augmented with domain-specific knowledge to move beyond hand-wavy answers. Retrieval-Augmented Generation (RAG), and more specifically its graph-based extension GraphRAG, is presented as one such solution.
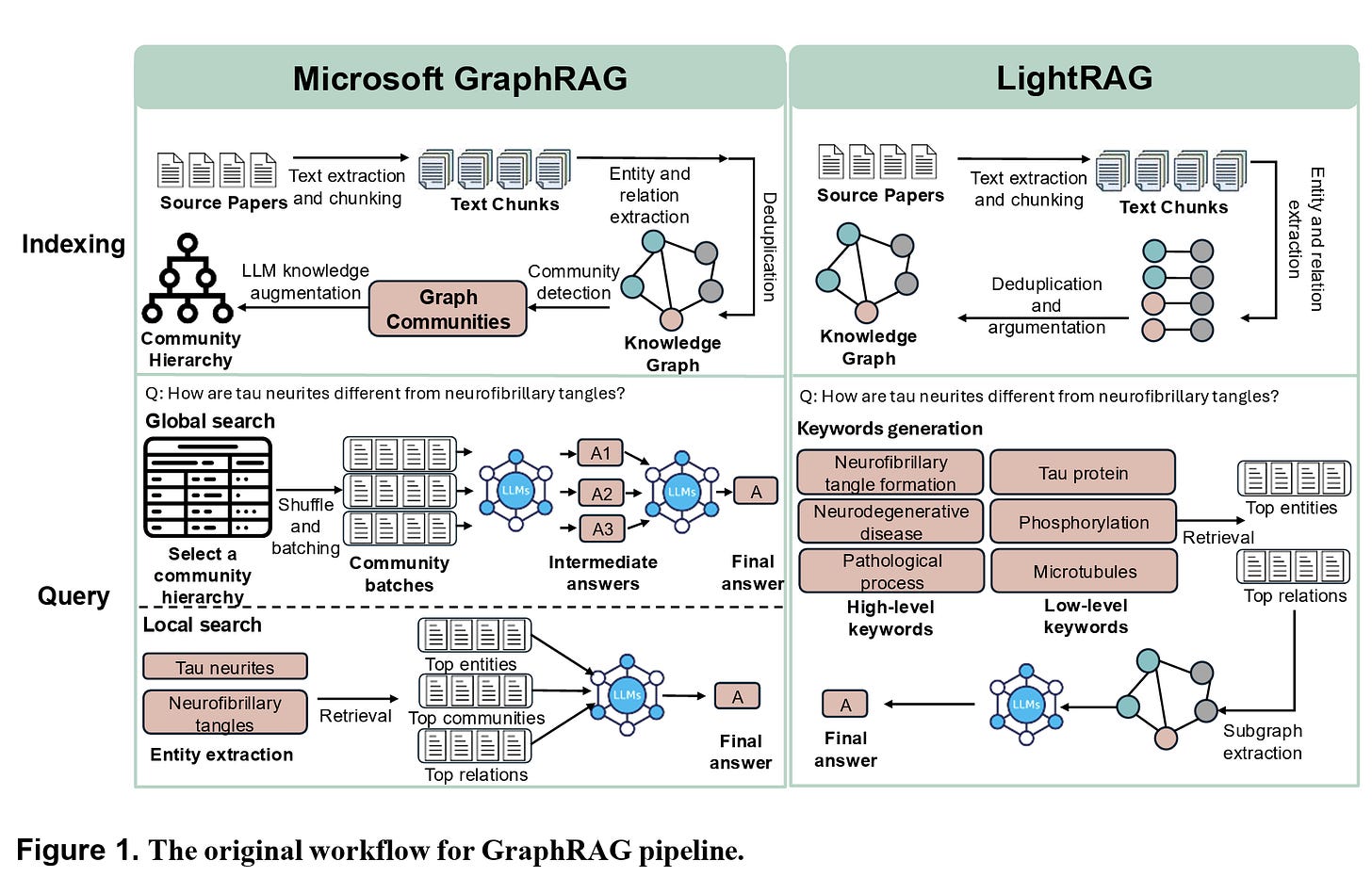
Figure 1 in the paper illustrates how these systems operate, showing the workflow of Microsoft GraphRAG and LightRAG side by side. Both aim to ground the model in curated scientific knowledge, but they do so in different ways,with Microsoft GraphRAG relying on hierarchical community structures and LightRAG opting for more direct keyword-driven retrieval.
At the heart of the paper lies a simple but powerful thesis: if we want LLMs to provide trustworthy scientific assistance, they need better context and better structure. GraphRAG builds knowledge graphs out of scientific papers, capturing not just terms but also relationships between them. Far from being a cosmetic tweak, the graph-based structure provides a way to retrieve information that is both relevant and organized.
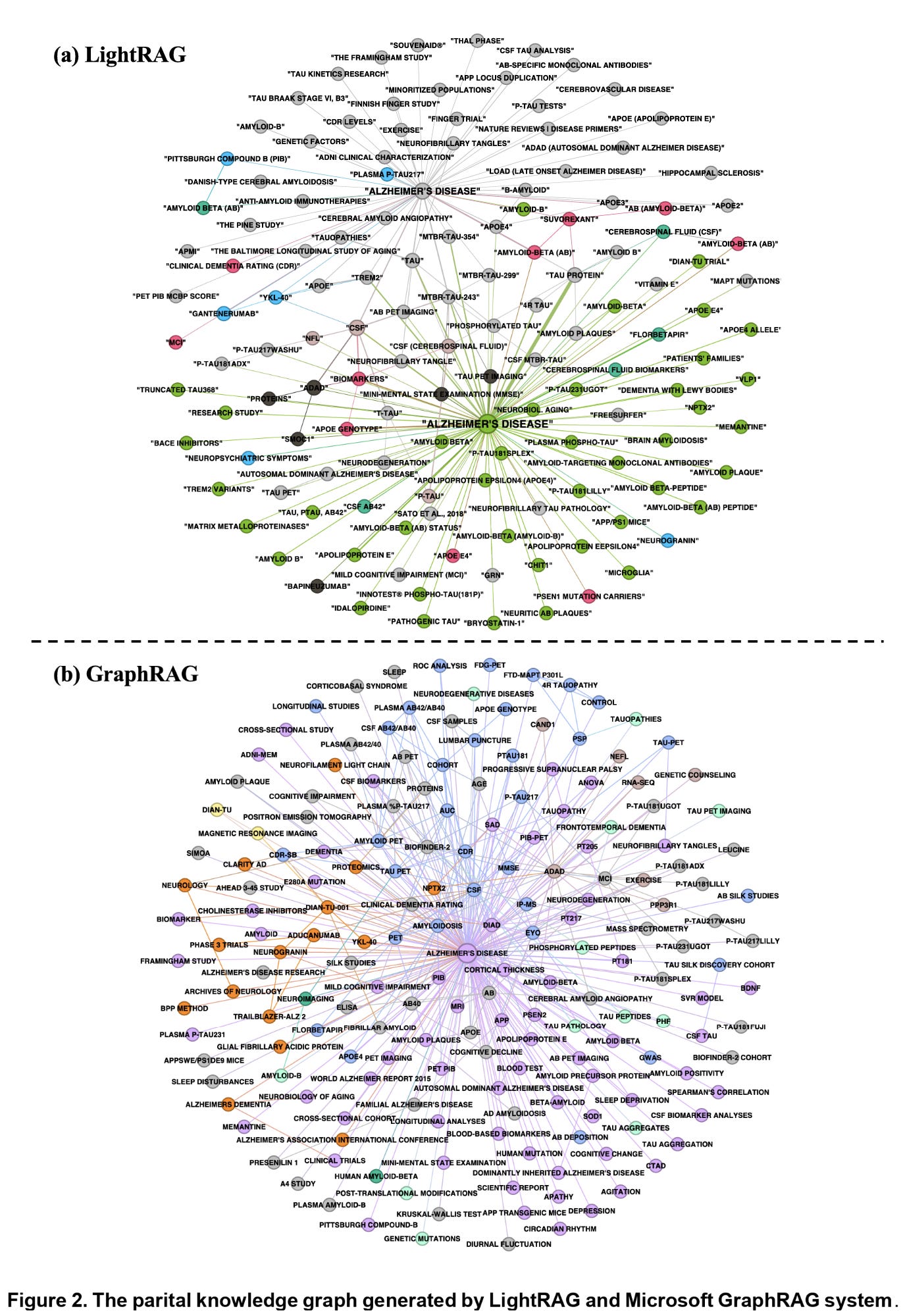
Figures 2 and 3 below help visualize this difference. Figure 2 shows the partial knowledge graphs generated by Microsoft GraphRAG and LightRAG, demonstrating how entities, relations, and concepts are connected.
Figure 3 then presents the evaluation prompts that the authors used to test for comprehensiveness, empowerment, diversity, and directness. Together, both figures highlight the dual goals of GraphRAG: to provide not only accurate answers, but answers that would allow a researcher to reason further.
The contrast between Microsoft GraphRAG and LightRAG is revealing. Microsoft’s system emphasizes community-level summaries and multi-hop reasoning across clusters of knowledge, while LightRAG focuses on more lightweight keyword extraction. The trade-off? Microsoft GraphRAG delivers richer answers but at a higher computational cost, while LightRAG is easier to update and extend but risks missing nuance if the wrong keywords are chosen.
The authors didn’t just stop at architectural descriptions. They conducted a systematic evaluation. They curated 50 Alzheimer’s disease papers and 70 expert-level questions spanning background, methods, results, and open-ended inquiry. Both Microsoft GraphRAG and LightRAG were benchmarked against a standard GPT-4o baseline.
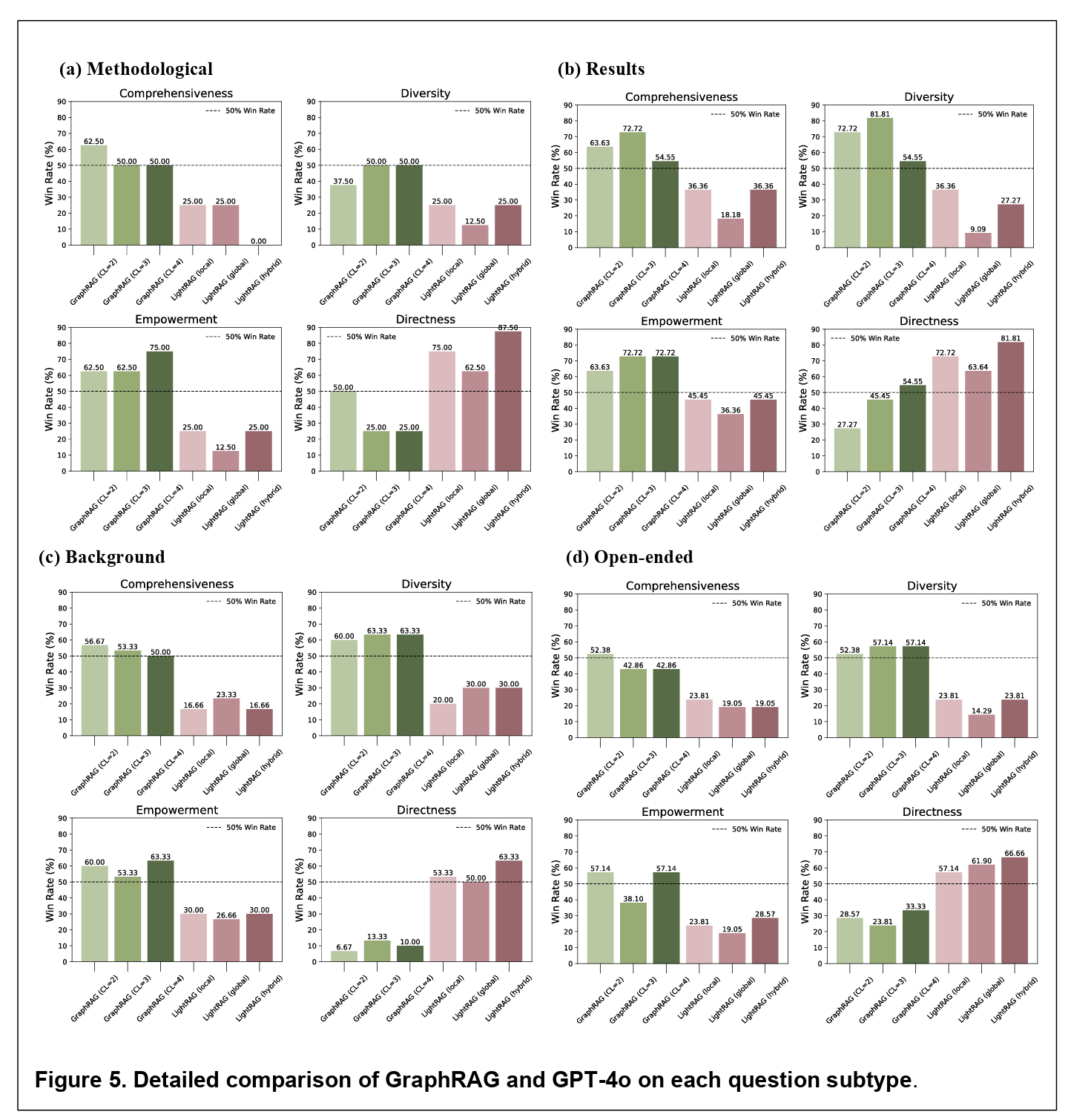
Figure 5 above provides a broad comparison of the models, breaking results down by question type. The findings are subtle and suggestive. Microsoft GraphRAG consistently outperformed GPT-4o in producing comprehensive and empowering answers, especially for result-specific questions. For example, when asked about which amyloid beta species have been measured using SILK, GraphRAG’s answer was not only more complete but also better organized. LightRAG, in contrast, fared worse on most dimensions, except for directness, where its more focused retrieval sometimes gave crisper replies.
Tables 1 and 2 below illustrate this with concrete examples. In Table 1, Microsoft GraphRAG identifies three specific amyloid beta species, while GPT-4o misses one. The difference is subtle but crucial. In biomedical science, omitting such a detail can mean the difference between a useful answer and a misleading one.
Yet the results also show limits. Standard GPT-4o remained surprisingly competitive in many cases. This suggests that as LLMs continue to evolve and incorporate more domain knowledge, the bar for specialized systems like GraphRAG will only get higher. And Microsoft GraphRAG’s reliance on exhaustive community iteration raises scalability concerns: what happens when the knowledge base expands from 50 papers to 50,000?
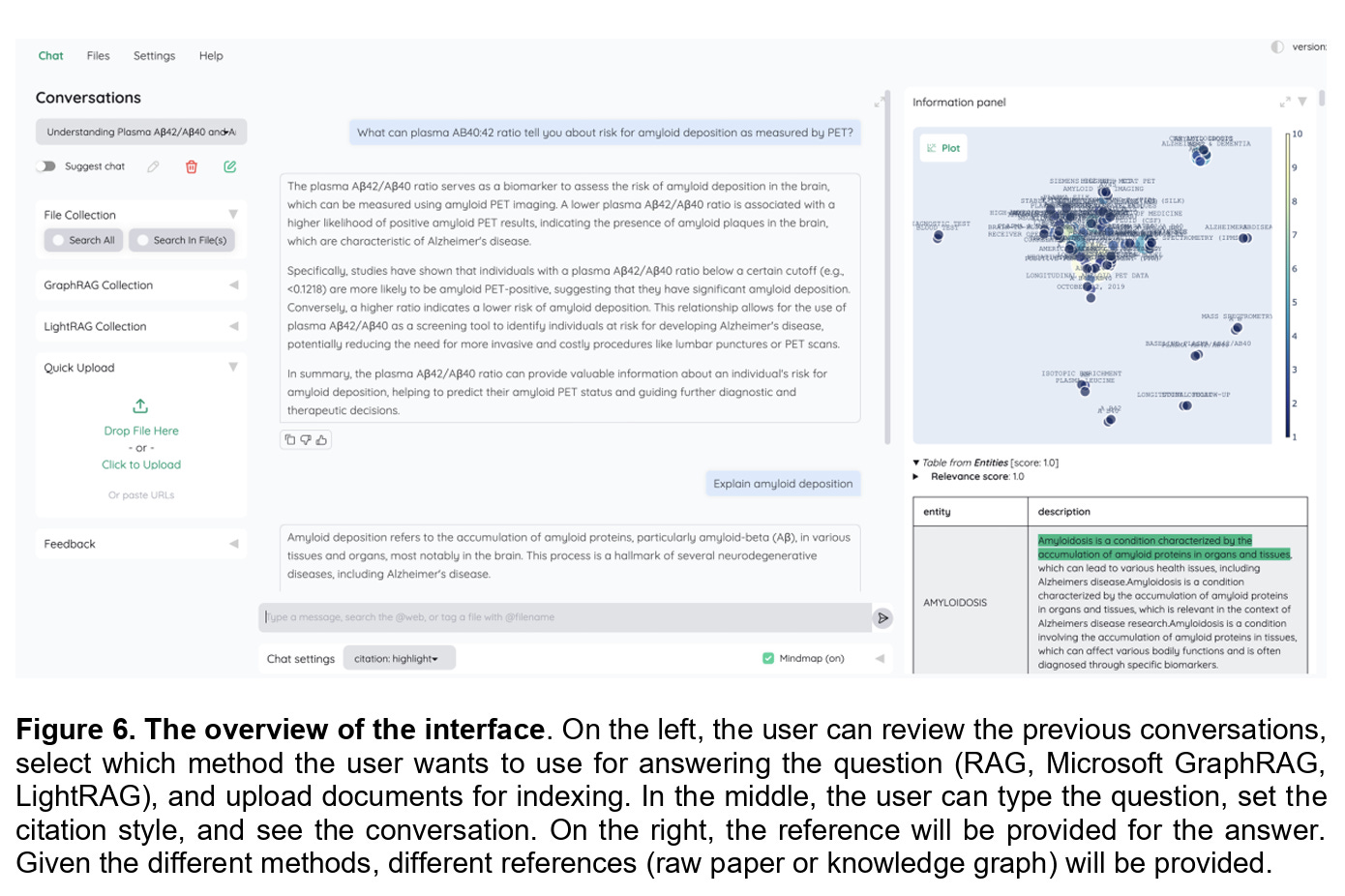
To make their findings practical, the authors went a step further and built a simple interface on top of an open-source repository. Shown in Figure 6, the interface allows researchers to choose between standard RAG, Microsoft GraphRAG, and LightRAG, and then query a pre-built Alzheimer’s disease database. The interface not only provides answers but also returns references in different formats depending on the method used. This makes the discussion of traceability tangible, moving it from theory into the hands of actual domain experts.
In closing the musing, it shows us that while LLMs are powerful, they are not enough on their own for biomedical research. Domain-specific augmentation, especially in the form of graph-based retrieval, can meaningfully improve both accuracy and usability. Second, it highlights the trade-offs that remain: Microsoft GraphRAG shines but at a cost, LightRAG is efficient but brittle, and standard LLMs are catching up faster than expected.
The broader message is that we are only at the beginning of figuring out how to responsibly deploy LLMs in scientific research. Accuracy is one hurdle, but traceability may prove even more important. Without it, researchers cannot trust the system enough to integrate it into serious workflows. With it, we might finally begin to see LLMs become real collaborators in scientific discovery.