Musing 53: Automatically Labeling $200B Life-Saving Datasets: A Large Clinical Trial Outcome Benchmark
An interesting benchmark paper out of UIUC
Today’s paper: Automatically Labeling $200B Life-Saving Datasets: A Large Clinical Trial Outcome Benchmark. Gao et al. 13 June. 2024. https://arxiv.org/pdf/2406.10292
Clinical trials are essential for testing the efficacy and safety of new drugs, but they are often hindered by high costs, lengthy durations, and high rates of failure due to issues such as low efficacy, safety concerns, and poor trial design. These challenges contribute to the escalating costs of drug development, which significantly impacts the overall efficiency of bringing new therapeutic options to market.
Despite the critical importance of clinical trials in the pharmaceutical industry, there is a surprising lack of readily accessible high-quality data on trial outcomes. Many trial results are not published, and those that are often remain disconnected from other trials or phases, making it difficult to track and predict outcomes comprehensively. This data gap presents a major obstacle for developing predictive models that could potentially streamline and enhance the drug development process.
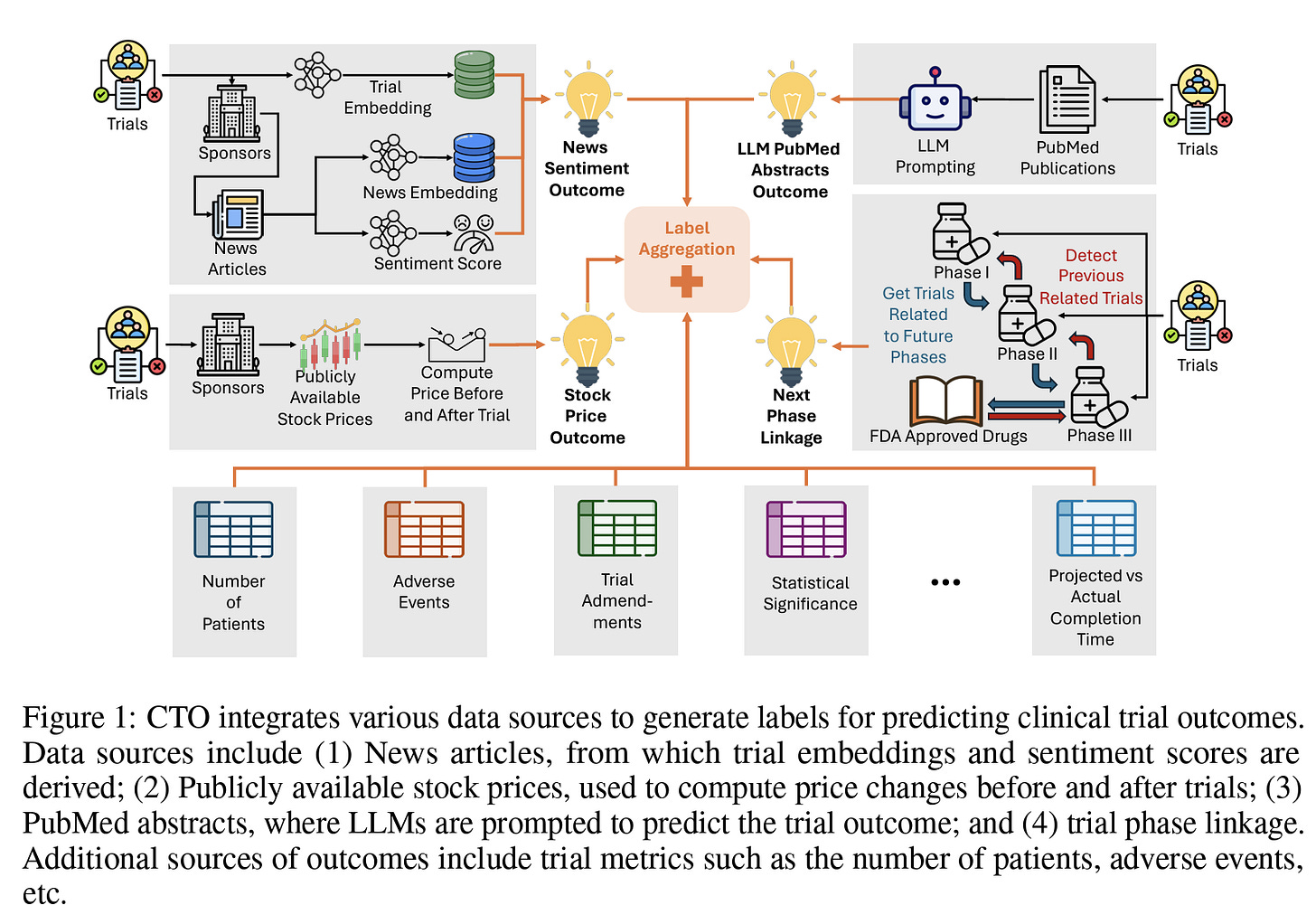
Today’s paper proposes the development of such predictive models using a large dataset of clinical trial outcomes, aiming to improve the prediction accuracy and help optimize the drug development pipeline. The authors propose CTO, the first large-scale, publicly available, open-sourced, and fully reproducible dataset of clinical trial outcomes derived from multiple sources of weakly supervised labels, including trial phase linkages, LLM interpretations of trial-related publications, news headline sentiments, stock prices of trial sponsors, and other trial metrics:
Previous studies have tackled clinical trial outcome prediction using various methods. Early work employed statistical analysis and ML models on limited private data sets (<500 samples). As shown below, all previous work relies on industry sponsors to obtain labels, and most are not easily accessible. The authors claim to be the first to aggregate publicly available data sources on a large number of trials:
CTO’s key methodology was outlined earlier in Fig. 1. PubMed abstracts have been automatically linked to trials by the Clinical Trials Transformation Initiative (CTTI) along with other initiatives. Initially, all PubMed abstracts for each trial were extracted using the NCBI API, and the statistics of these abstracts are detailed in the supplementary materials. These abstracts are classified into categories: Background, Derived, and Results. Focusing on clinical trial outcomes, only abstracts categorized as Derived and Results were utilized. For trials that had multiple abstracts, the top two abstracts were selected based on their title similarity to the official title of the trial to ensure relevance. Using these selected abstracts, the 'gpt-3.5-turbo' model was employed to summarize crucial trial-related statistical tests and predict outcomes. Moreover, this large language model was also used to generate question-answer pairs about the trial.
The journey of a drug from discovery to FDA approval involves several stages, beginning with Phase 1 trials to assess safety and dosage. Subsequent Phase 2 and 3 trials evaluate efficacy and compare the new drug to existing therapies. Upon completing Phase 3, a drug may be submitted for FDA approval. A key limitation of the CTTI dataset is the lack of connectivity between trial phases, which could significantly enhance the ability to analyze trial progression and outcomes based on advancement to subsequent phases. Moreover, linking trials across phases is not straightforward due to challenges, including unstructured data, inconsistent reporting standards, missing information, data noise, and discrepancies in intervention details across phases. Despite these challenges, linking trials can be invaluable, particularly as a source of weak labels in clinical trial outcome prediction tasks. The authors propose a novel trial-linking algorithm, which, to their knowledge, is the first attempt to systematically connect different phases of clinical trials. The trial linkage extraction process consists of two primary steps: 1) Linking trials across different phases, as illustrated below in Figure 2B, and 2) Matching phase 3 trials with FDA approvals, as shown in Figure 2C.
In total, the authors end up with more than 450k trials with automatically labeled outcomes. In experiments, they compare their aggregated labels, sourced from various weak labeling methods, against the TOP test set, which is annotated by human experts. The primary goal is to assess the agreement between their labels and the human-annotated labels. The results suggest that there is, although determining the better labeling method is subjective and depends on various factors (see below). Specifically, the figure illustrates the distribution of labels generated by the random forest and data programming labeling functions, showing a significant distribution difference from that of Data Programming (RF predicts almost all trials to be successful). This discrepancy can be attributed to the nature of the random forest labeling function, which relies on a small-scale, human-annotated set that might not capture the overall trial distribution. However, this leads to better agreement with the TOP test set than with data programming.
On the other hand, data programming is an unsupervised approach that generates labels by considering the agreement and relationships between all weak labeling functions across the entire dataset. This approach allows data programming to more accurately learn the trial outcome label distribution. Due to this challenge, the authors provide both labels from the random forests and the data programming labeling function in our dataset release. Additionally, Figure 4 A and B above illustrate the distribution of labels across phases and trial statuses in the proposed CTO dataset. This highlights the sheer quantity of the dataset compared to previous methods.
In closing, the dataset and labels facilitate the development of prediction models not only for drug interventions but also for biologics and medical devices, broadening the scope of clinical trial outcome research. The models consistently perform comparably to baseline models across all phases and metrics when benchmarked against TOP, underscoring the effectiveness of weak supervision and the reliability of the methodology. The authors recognize that automatically created labels will never be a substitute for human ones. However, for data-hungry new ML methods in clinical trial optimization, they assert that this could be a good first step before obtaining human labels, due to the high agreement. In addition, open-source availability means that any customization to specific tasks can be made quickly and reproducible. The dataset is made available at https://github.com/chufangao/CTOD.
I was also very impressed with the nearly-20 page appendix and datasheet that came with the paper. This is a general trend I’m seeing with good papers (more details), especially since several conferences now allow unlimited appendices. Even if they don’t, an extended version can always be published on arxiv, or a link can be provided as supplementary material. I look forward to exploring this dataset further.





Thank you, Mayank, for sharing our benchmark paper and for writing such a holistic article about it!