Musing 87: Can Large Language Models Replace Data Scientists in Clinical Research?
A provocatively titled paper out of Kyoto, Osaka and UIUC
Today’s paper: Can Large Language Models Replace Data Scientists in Clinical Research? Wang et al. https://arxiv.org/pdf/2410.21591. 28 Oct. 2024
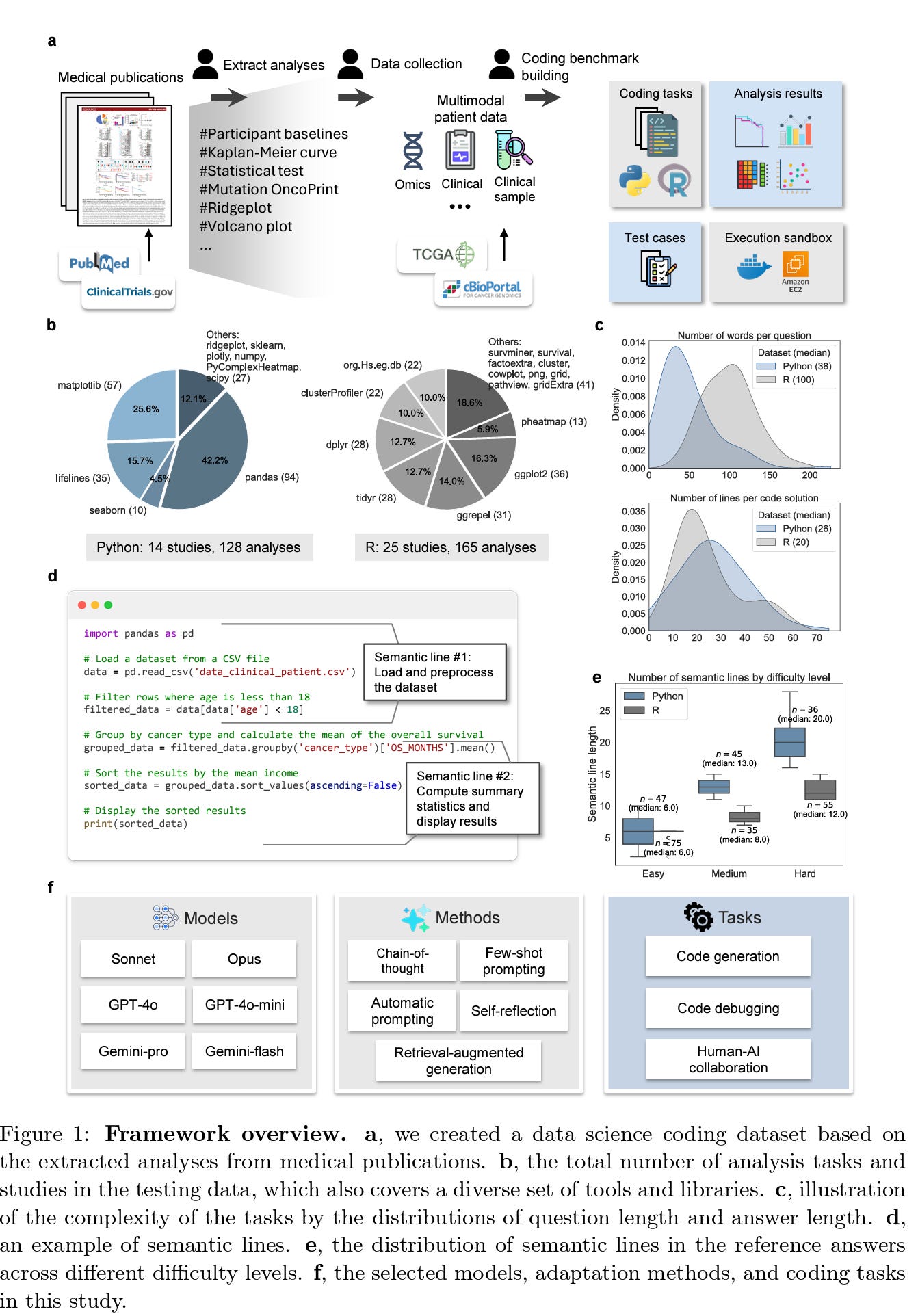
Data science plays a critical role in clinical research, but it requires professionals with expertise in coding and medical data analysis. In today’s brief musing, I describe how the authors develop a dataset consisting of 293 real-world data science coding tasks, based on 39 published clinical studies, covering 128 tasks in Python and 165 tasks in R. This dataset simulates realistic clinical research scenarios using patient data. The process is captured below in a nice figure.
Based on the figure, 1a outlines the process of dataset creation by extracting analyses from medical publications, resulting in a range of coding tasks focused on clinical research scenarios, such as Kaplan-Meier curves and mutational OncoPrints. It proceeds from there and is fairly self-explanatory. 1d is very interesting as it describes “semantic lines,” a metric used to determine task complexity by counting lines of code performing unique operations. 1e presents the distribution of these semantic lines across task difficulty levels, showing that Python tasks are typically more complex than R tasks due to differing language capabilities (I have to confess this is news to me; I always considered R to be more complex for some reason).
For evaluation, the authors assessed the combinations of models and adaptation methods across three tasks: code generation, code debugging, and human-AI collaboration. The first two tasks were used to measure the models’ accuracy in automating the coding process. They employed Pass@k as the primary metric, where k represents the number of attempts the model is allowed to make to solve a coding task. This metric behaves like the probability that at least one out of the k attempts is correct. Specifically, they selected k = 1 as a strict benchmark to evaluate how well LLMs can automate tasks on the first attempt, providing insight into their immediate accuracy. Additionally, they used k = 5 as a more relaxed metric to explore the model’s potential to improve given multiple attempts, offering a broader assessment of its ability to generate correct solutions when allowed more tries.
Figure 2 below describes assessment of different models and adaptation methods. For each task, the LLM is provided with a raw question that describes the target task, as well as a dataset description (Fig. 2a). The description contains spreadsheet names, column names, and common cell values, which guide the LLM in identifying the correct spreadsheet, column, and values to work with. The results show that current LLMs cannot consistently produce perfect code for clinical research data science tasks across all difficulty levels (2d). For Python tasks, the Pass@1 scores vary significantly based on task difficulty. For Easy tasks, most LLMs achieve Pass@1 rates in the range of 0.40-0.80. However, for Medium tasks, the Pass@1 rates drop to 0.15-0.40, and for Hard tasks, they range from 0.05 to 0.15. Performance differences also exist between different LLMs, particularly within the same series.
Adjusting the temperature settings, the authors sampled multiple solutions from LLMs and calculated Pass@5 scores for Python tasks (2b). In most cases, increasing the temperature allows LLMs to generate more creative and diverse solutions, resulting in higher probabilities of producing a correct solution. This trend was consistent across all models, suggesting the potential benefit of having LLMs brainstorm multiple solutions to reach better outcomes.
Motivated by the varied performances of LLMs with different instruction levels, the authors further hypothesized that tailored adaptations for LLMs in clinical research data science could lead to greater improvements. To test this, they introduced two key dimensions of adaptation: (1) enhancing LLM inference and reasoning by incorporating advanced instructions or external knowledge, and (2) employing multiple rounds of trial-and-error, allowing LLMs to iteratively correct their errors. The results of these adaptations are shown in Fig. 3 below.
Studying the figure, comparison of adaptation strategies based on GPT-4o is illustrated in 3b. Each data point represents the average Pass@1 score achieved for coding tasks in a given study. A point on the diagonal line indicates equivalent performance between the adaptation and the vanilla method. Studying the results, they find that AutoPrompt overfitted on the training tasks and struggled to generalize effectively on the testing tasks, while RAG performed similarly to Vanilla, despite incorporating external knowledge into the inputs. The authors hypothesize this is because GPT-4o was likely trained on a wide range of public sources, including package documentation, webpages, medical articles, and online guidelines.
ManualPrompt provided a modest improvement, boosting Pass@1 by an average of 10% across studies and outperforming Vanilla in 7 out of 11 cases. This demonstrates the effectiveness of incorporating expert knowledge to better adapt LLMs to specific tasks.
Moving on to the user study conducted by the authors in Figure 4 below, the authors first developed a platform that integrates LLMs into data science projects for clinical research. The interface supports real-time interactions, allowing users to generate and execute code in a sandbox environment with instant visualizations. In the study itself, the authors recruited five medical doctors with varying levels of coding expertise. Each participant was assigned three studies, with approximately 10 coding tasks per study (4d). Users worked with LLMs on the platform to complete these tasks and submitted their solutions once their code passed all the test cases.
The results of the code comparison analysis are presented in 4c, showing the distribution of the proportion of user-submitted code derived from LLM-generated solutions. The authors found that a significant portion of the user-submitted code was drawn from AI-generated code. For Easy tasks, the median proportions were 0.88, 0.87, and 0.84 across the three studies, indicating that users heavily relied on LLM-provided solutions when crafting their final submissions. For Medium and Hard tasks, the ratios were generally lower: in Study 1, the proportions were 0.44 for Medium tasks and 0.96 for Hard tasks, while in Study 2, the proportions were 0.75 for Medium and 0.28 for Hard tasks. These findings demonstrate the potential of LLMs to streamline the data science process, even for users without advanced coding expertise, with greater reliance on LLMs for easier tasks and more mixed results for more complex ones.
Quantitative results from the user survey are shown in 4e. The average user ratings for each category are: Output Quality (3.4), Support & Integration (3.0), System Complexity (3.5), and System Usability (4.0). These ratings suggest that, overall, users had a positive experience using the platform, although there is room for improvement.
In closing the musing, I thought that the title of the paper was a little too provocative, but the figures were extremely well designed; so much so, that reading the paper itself is minimally required. The figures are self-explanatory and clean. I wish figures that I have drawn for my own papers were this good.
Two critical findings from our experiments are: (1) When human experts provide more detailed, step-by-step instructions, the quality of LLM-generated code significantly improves, as demonstrated by the superior performance of Chain-of-Thought (CoT) prompting (Fig. 3b earlier). (2) Although LLM-generated code is often imperfect, it serves as a strong starting point for human experts to refine. This confirms what many of us have found in our daily lives; that LLMs are better served as copilots for humans, not human replacements.
There are also some supplementary results and figures. Figures aside, the results show that LLMs cannot yet replace data scientists in clinical domains, at least not the scientists who are really good at their job and capable of handling complex tasks. But perhaps the writing is on the wall!